Lab 11: Pre-Lab
Over the next several years BIOL 121 students will be testing meat samples from Kenya, through DNA analysis, to determine if poaching and bushmeat use is threatening conservation efforts. Due to the new "online only" nature of this lab, you will be provided the output of various steps so you can complete the process of species identification without doing any bench work. In labs 10-12, your task will be to identify the species of origin of a meat samples from Kenyan butcheries. You will learn about poaching, the bushmeat crisis and practice key techniques to complete DNA analysis of your samples. To prepare for Lab 11, please review this pre-lab. Once you feel confident regarding the below topics, and have your Lab Notebook ready from last week, complete the corresponding LABridge in Blackboard.
-
Introduction/Review
-
Do you know enough?
-
What would we have done in lab?
-
LABridge
<
>
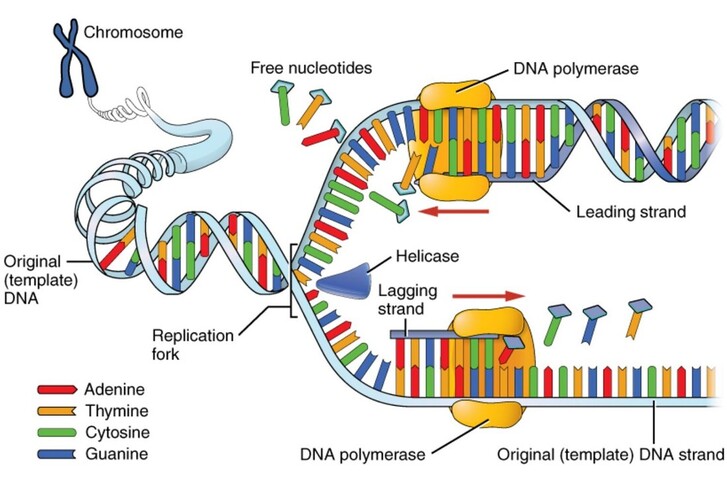
Review the Steps of DNA Analysis
In our last lab, we reviewed steps 1 and 2. In Lab 11 we will focus on Polymerase Chain Reaction (PCR).

Please review the steps and sequence below, with a focus on step 3.
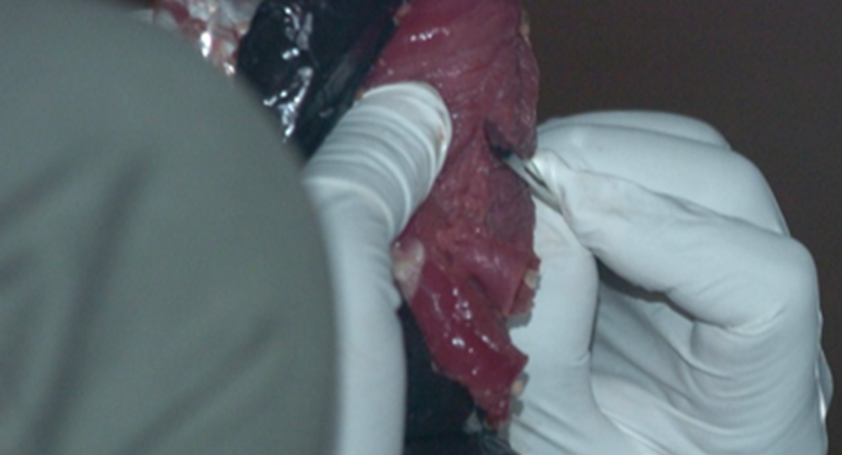
1) Sample PROCESSING
Bushmeat Processing: Using aseptic techniques, the bushmeat samples are cut into approximately 1 cc sections. They labeled and stored in ethanol at -20 degrees Celsius. Special care is taken to ensure no cross-contamination or human contamination occurs. Samples are then carried back or shipped to the WKU biotechnology Center. -----> This has already been done. |
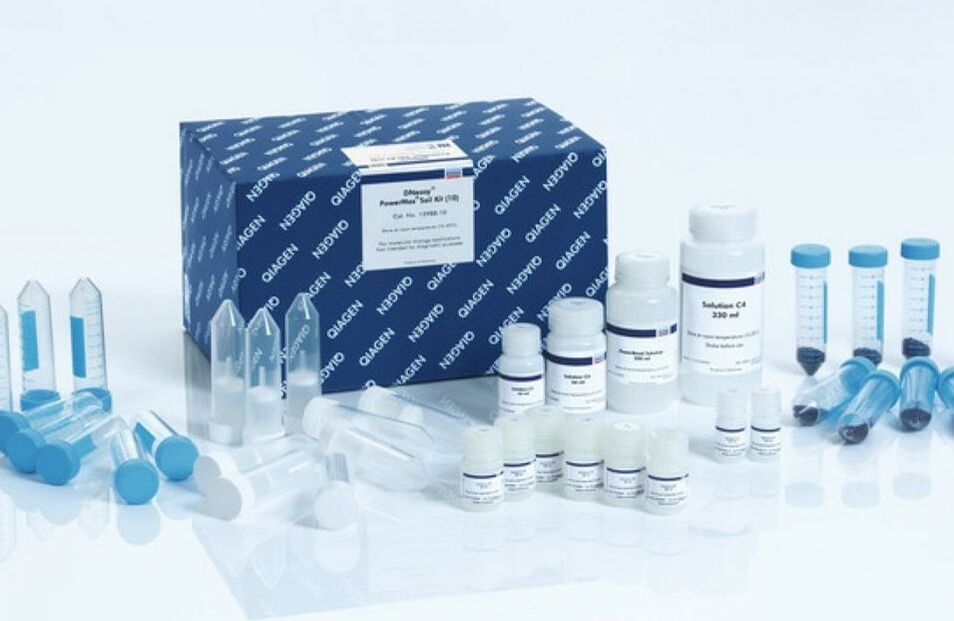
2) Digestion & Extraction
Digestion liquefies the tissue in such a way that keeps the DNA intact for extraction. We use special "kits" as pictured to streamline the process. Extraction requires more steps to break through the cell and organelle membranes to free the DNA. Once extraction is complete, the DNA sample is tested to ensure an adequate amount of intact DNA was extracted from the sample. -----> This process has already been done with our meat samples. In Lab 10, you will practice extracting DNA from a strawberry to better understand this process |
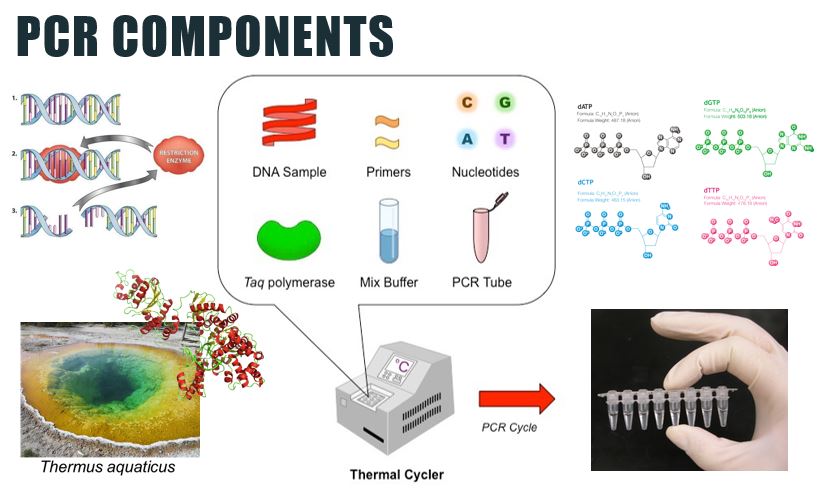
3) Polymerase chain reaction (PCR)
PCR makes copies of a DNA fragment from one original copy. The goal is to amplify a specific region, the target DNA or gene of interest (GoI), depending on the type or goal of research. The PCR cocktail includes the following ingredients: the DNA sample, primers (short sequences of RNA or DNA that start replication), dNTPs (free nucleotides), taq polymerase (a heat stable form of DNA polymerase derived from bacteria) and a buffer solution. There are three steps to PCR in which the temperature is cycled (in the thermo"cyler"). You need to know the steps and what happens in each! The total number of resulting DNA strands is (the number of original strands) X 2^n, where n = the number of PCR cycles. -----> In Lab 11, you will be completing PCR on the extraction products of our bushmeat samples and asked make some calculations. |
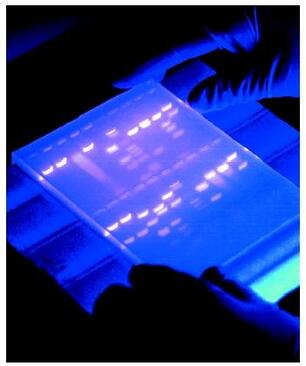
4) Gel electrophoresis
Agarose gel electrophoresis is a method used to separate DNA strands by size, and to determine the size of the separated strands by comparison to strands of known length. Your PCR products are deposited in the top of the gel. Using electricity, the DNA (with a negative charge) is pushed through the gel towards the positive electrode. As your gel "runs," the DNA is separated by size. The DNA strands show up as bands under UV light and you can read the results. Your products can be compared with the ladder or marker, which has standard sized DNA fragments of KNOWN length used for comparison. In this way, you can know the exact length of your DNA samples. -----> You will MAKE & RUN an agorose gel in Lab 11 to make sure our PCR product contains the cytochrome b gene. |
5) Sequencing
Once we know we have amplified (copied) the right gene we are ready to sequence the gene. We expect the sequence (the order of As, Ts, Cs and Gs) within the cytochrome b gene to be different for different species. Samples are placed into a sequencer apparatus which can detect the order of nucleotide bases in our sample. The sequence is then cleaned and edited, |
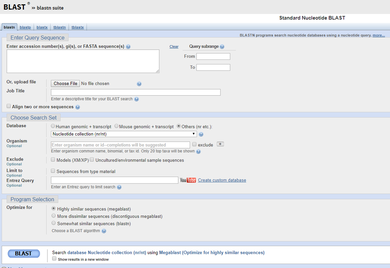
6) BLASTing
6) BLASTing: The National Institute of Health (NIH) and National Center for Biotechnology Information (NCBI) hosts a database called GenBank, which houses all known DNA sequences. Once the sequences of our samples are ready, they are pasted into a search tool (called a BLAST) which matches them to the correct species! -----> You will be provided the sequence of successful samples in Lab 11 and asked to determine the species of origin in lab. |
PCR Uses & Ingredients
Once DNA has been extracted, it is mixed into a particular PCR solution containing:
- Taq polymerase: A type of heat-stable DNA polymerase derived from a species of bacteria living in hot springs. Because Taq polymerase continues to function normally at high temperatures, using it allows researchers to separate the DNA strands without destroying the polymerase.
- Primers: Short, single-stranded sequence of RNA or DNA that enables the start of replication of a DNA sequence that is synthesized from the 3’ end of the primer. Two types are needed, a forward and a reverse primer.
- Deoxynucleoside triphosphates (dNTPs): Free nucleotides to be used in constructing the new copies of DNA.
- Mix buffer: Necessary to create optimal conditions for activity of Taq DNA polymerase and may contain restriction enzymes, which act like molecular scissors cutting the copied DNA strands at particular locations based on their genetic code.

Learn the ingredients (components) required for PCR to work. You also need to know what each one is and why it is required!
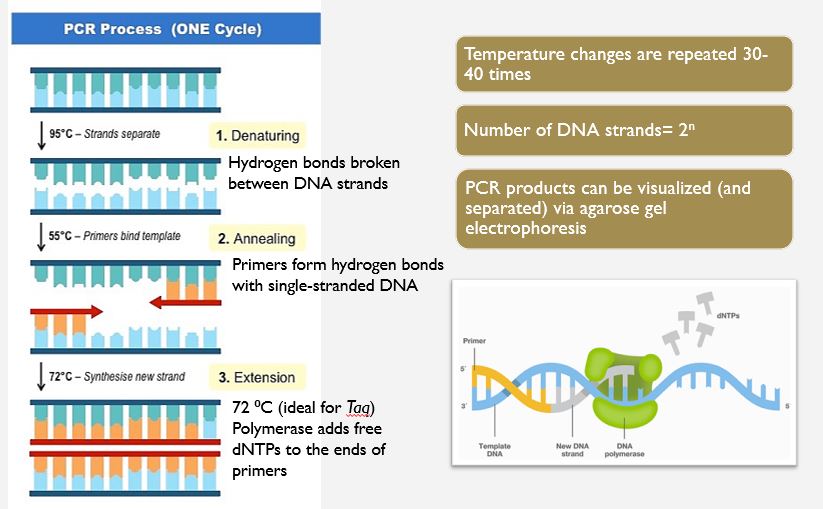
The Steps of PCR
We would have created PCR cocktails for each of our bushmeat samples and placed into a thermocycler for synthesis. Instead, you will do a bit of research and a virtual PCR lab. Then you will be provided with the PCR outcomes to complete some analysis.
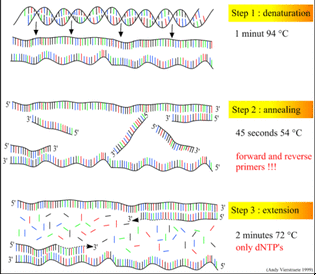
The PCR mixture is placed inside a thermocycler (PCR machine). It is typically repeated about 35 times and the temperature changes are programmed by researchers and automated by the thermocycler. The process proceeds in three steps as outlined below.
- Denaturation: the solution is first heated to nearly boiling—95ºC. The heat breaks the hydrogen bonds between the two DNA strands and allows them to separate.
- Annealing: the temperature is dropped to around 60ºC. The exact temperature depends on the length and base composition of the primers. At this relatively low temperature, the primers can form hydrogen bonds with the single-stranded DNA. Two primer types are created, each one complementary in sequence to one of the two ends of the target DNA. To make the primers, the sequences at the ends of the target DNA must be known.
- Extension: the temperature is increased to 72ºC. This is the optimal temperature at which Taq polymerase functions. The primers are essential in this process, because they provide free 3’ hydroxyl groups, to which the polymerase can add additional dNTPs. Each new dNTP that joins the growing strand is complementary to the nucleotide in the opposite strand.

You need to know the three steps of PCR and should be able to describe what happens in each step. You also need to know how we determine the results of PCR...how can we know how many copies we have after the procedure?
At the end of this first cycle, there are two DNA copies instead of the one original copy. The process continues, doubling the number of target DNA copies with each cycle.
The general formula for the number of DNA strands created by PCR is
N2^n
N = the number of original strands
n = the # of PCR cycles.
The general formula for the number of DNA strands created by PCR is
N2^n
N = the number of original strands
n = the # of PCR cycles.
-
Exercise I
-
Exercise II
-
Exercise III
<
>
Exercise I. Compare and Contrast:
|
|
|
|
Exercise II. Virtual PCR
|
|
Exercise III. Determine PCR Yield
|
Procedure
|
At the end of this first cycle, there are two DNA copies instead of the one original copy. The process continues, doubling the number of target DNA copies with each cycle. The general formula for the number of DNA strands created by PCR is N(2^n) N = the number of original strands n = the # of PCR cycles. |
|
Lab 11 BIOL 120 CONNECTIONS Section 1.6: Doing Biology Big Picture 1: How to Think Like a Scientist Chapter 4: Nucleic Acids Chapter 8: Enzymes Chapter 12: The Cell Cycle Chapter 15: DNA and the Gene: Synthesis & Repair Chapter 16: How Genes Work Chapter 20: The Molecular Revolution Chapter 54: Biodiversity and Conservation Biology *BIOL 122 |
KAS citation format:
Mountjoy, N.J 2021. Title of page. Biological Concepts: Cells, Metabolism & Genetics. https://www.121cellmetagen.com. Date accessed (MM/DD/YYY).
Mountjoy, N.J 2021. Title of page. Biological Concepts: Cells, Metabolism & Genetics. https://www.121cellmetagen.com. Date accessed (MM/DD/YYY).