Lab 10: Pre-Lab
Over the next several years BIOL 121 students will be testing meat samples from Kenya, through DNA analysis, to determine if poaching and bushmeat use is threatening conservation efforts. We are currently testing the procedures that will be used for this research project. You will be provided the output of various steps so you can complete the process of species identification. In labs 10-12, your task will be to identify the species of origin of a meat samples from Kenyan butcheries. You will learn about poaching, the bushmeat crisis and practice key techniques to complete DNA analysis of your samples. To prepare for Lab 10, please review this pre-lab. Once you feel confident regarding the below topics, and have your Lab Notebook ready from last week, complete the corresponding LABridge in Blackboard.
-
Introduction/Review
-
Do you know enough?
-
What would we have done in lab?
-
LABridge
<
>
What is bushmeat?
Kenya's wildlife is in decline in part due to poaching of commercially valuable species. In many areas, poaching in the form of snaring is commonplace, largely due to a lack of resources, food insecurity and poverty. Increased poaching effort has reportedly led to an increase in bushmeat in Kenya's markets and butcheries.

Watch the brief clip below about the Kirk's Dik Dik, a commonly snared species in our research area.
|
|
The dik dik (Madoqua sp) is likely the most commonly snared wildlife species in the Tsavo area. They are amazing little antelopes. There are four recognized species. Dik dik mate for life and are always in pairs, so if one is snared and killed, the other often dies as well. This YouTube video was the best footage I could find on them, but it is NOT an endorsement of "Rob the Ranger." |
Where did our meat samples originate? |
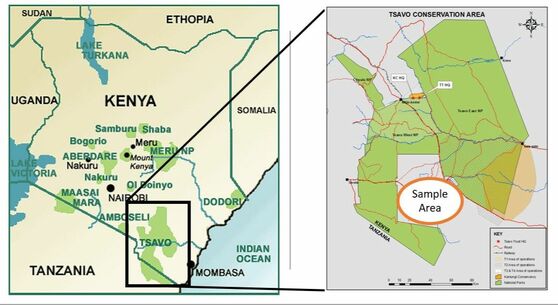
Our samples are from the Taita Taveta district of southeastern Kenya, which includes Kenya's largest national park system, Tsavo East and Tsavo West National Parks. Specifically, our samples are from the Kasigau area between the two parks, located on the trailing edge of the Eastern Arc Mountains. The Kasigau landscape is dominated by Mt. Kasigau, which the Titata people settled around to serve as a water catchment. The area also serves as a migration corridor between Tsavo East and West National Parks and is rife with human wildlife conflict.
|
Over the next several years BIOL 121 students will be testing samples from the five villages that surround Mt. Kasigau: Mwakasinyi, Keteghe, Rukanga, Jora and Bungule. Every semester we will be adding to a bushmeat database, which can be used by conservation groups and the Kenyan Wildife Service to locate hotspots of poaching and bushmeat activity.
This semester's samples were sourced from 8 butcheries (view slideshow) from the villages of Rukanga (samples 1-6), Jora (sample 7) and Bungule (sample 8).
This semester's samples were sourced from 8 butcheries (view slideshow) from the villages of Rukanga (samples 1-6), Jora (sample 7) and Bungule (sample 8).

View the slideshow below. It pictures our test butcheries as well as provides their GPS coordinates. Make sure you know from which villages are samples are sourced for this round of research.
3) What will we do in lab & how will we do it?
You might be wondering...why do we need DNA analysis to identify meat samples? As you saw in the introduction to this lab, meat sold in rural Kenyan butcheries does not look like meat sold in most places in the US. It is largely unregulated and unlabeled. Once meat is chopped up for sale, it is impossible to tell if it came from a cow, goat, or impala or elephant. In this way, poachers can sell their wildlife meat as domesticated meat products to the public.
Over the next several years BIOL 121 students will be testing meat samples from Kenya, through DNA analysis, to determine if poaching and bushmeat use is threatening conservation efforts. In labs 10-12, your task will be to identify the species of origin of a meat samples from Kenyan butcheries.
This week you will:
DNA extraction includes the use of an extraction buffer and ethanol:
Over the next several years BIOL 121 students will be testing meat samples from Kenya, through DNA analysis, to determine if poaching and bushmeat use is threatening conservation efforts. In labs 10-12, your task will be to identify the species of origin of a meat samples from Kenyan butcheries.
This week you will:
- Learn about DNA extraction by extracting DNA from fruit samples.
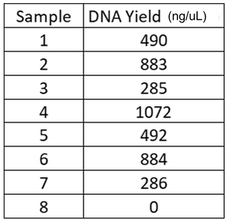
- Be given the extraction results of 8 bushmeat samples and asked to determine the quantity of product we may have.
- Review Polymerase Chain reaction and complete a short virtual lab.
DNA extraction includes the use of an extraction buffer and ethanol:
- Our extraction buffer includes detergent to break apart lipids and proteins and "free" the DNA from the nucleus (to get it out of the cell).
- It also includes salt with (+) charged sodium ions to neutralizes the (-) charge on the sugar-phosphate backbone of DNA, making it less soluble in water (so it stays together).
- We will use ethanol to separate the DNA from the mixture. DNA is not soluble in ethanol so it is visible (so we can see it).

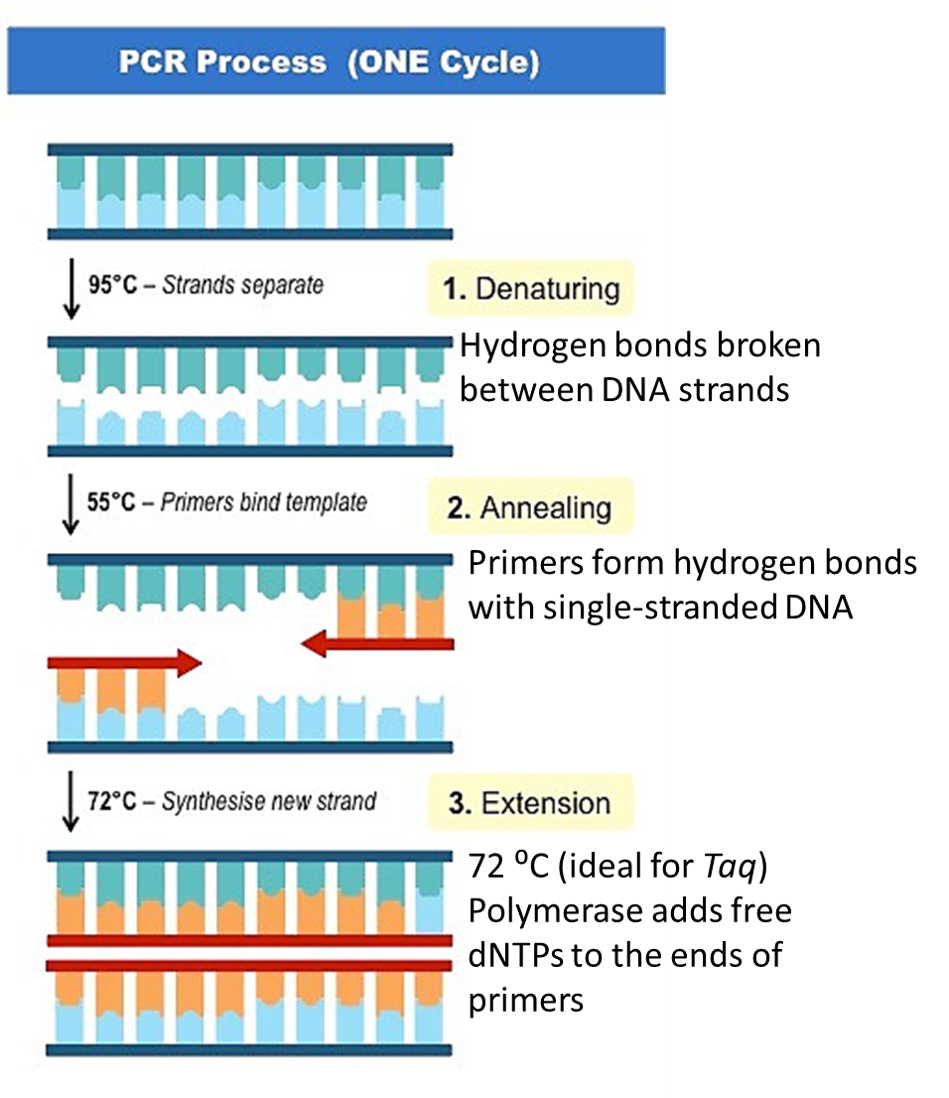
Read through the steps below. Pay careful attention to the details of steps 1, 2 and 3, and be sure you know the order of all six!
You will see these steps quite a few times. Start learning them now! Steps 1 -3 are the focus of lab this week! We will look again at step 3 next week and step 4. We will finish our analysis in Lab 12 with steps 5 and 6.
1) Sample PROCESSING
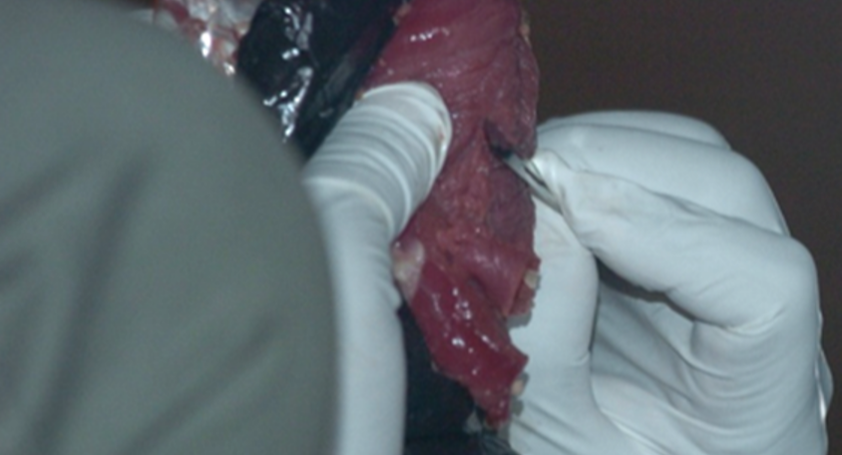
Bushmeat Processing: Using aseptic techniques, the bushmeat samples are cut into approximately 1 cc sections. They labeled and stored in ethanol at -20 degrees Celsius. Special care is taken to ensure no cross-contamination or human contamination occurs. Samples are then carried back or shipped to the WKU biotechnology Center. -----> This has already been done. |
2) Digestion & Extraction
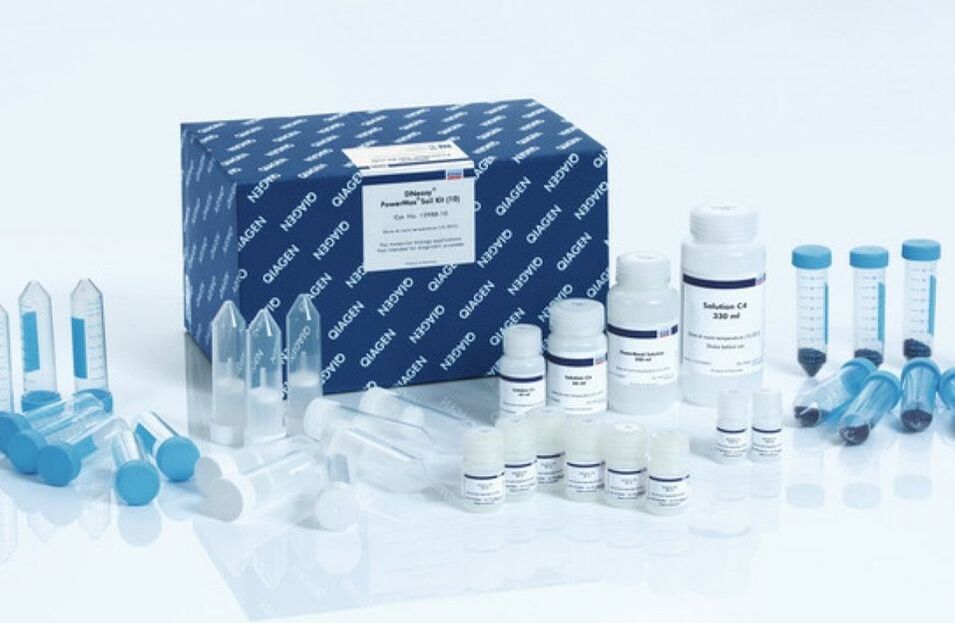
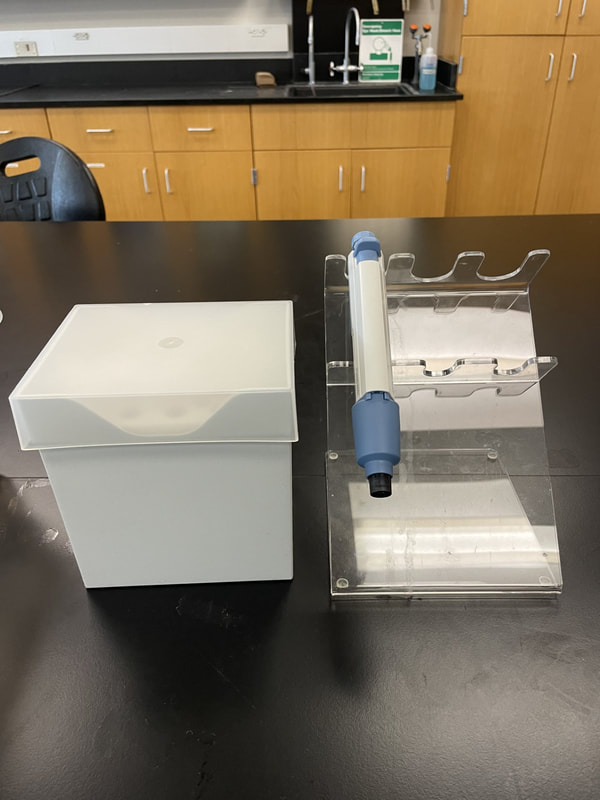
Digestion liquefies the tissue in such a way that keeps the DNA intact for extraction. We use special "kits" as pictured to streamline the process. Extraction requires more steps to break through the cell and organelle membranes to free the DNA. Once extraction is complete, the DNA sample is tested to ensure an adequate amount of intact DNA was extracted from the sample. -----> This process has already been done with our meat samples. In Lab 10, you will practice extracting DNA from your own cheek cells. |
3) Polymerase chain reaction (PCR)
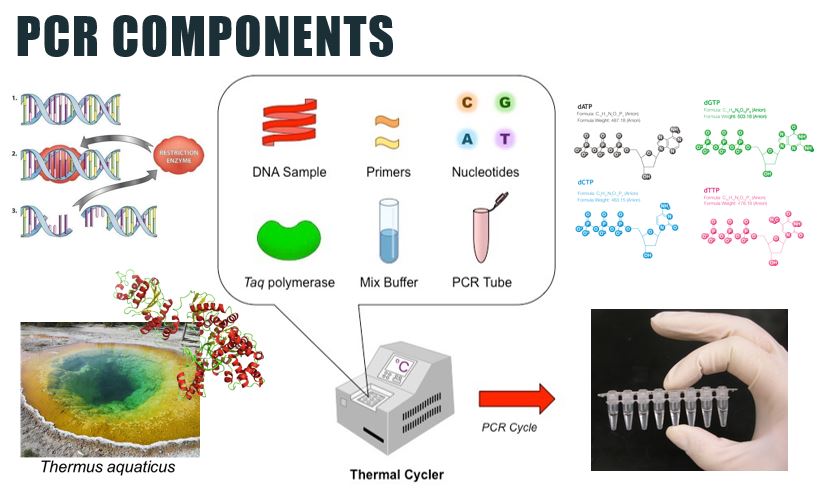
PCR makes copies of a DNA fragment from one original copy. The goal is to amplify a specific region, the target DNA or gene of interest (GoI), depending on the type or goal of research. The PCR cocktail includes the following ingredients: the DNA sample, primers (short sequences of RNA or DNA that start replication), dNTPs (free nucleotides), taq polymerase (a heat stable form of DNA polymerase derived from bacteria) and a buffer solution. There are three steps to PCR in which the temperature is cycled (in the thermo"cyler"). You need to know the steps and what happens in each! The total number of resulting DNA strands is (the number of original strands) X 2^n, where n = the number of PCR cycles. -----> In Lab 10, you will be given PCR products from our meat samples in lab and asked make some calculations. |
4) Gel electrophoresis
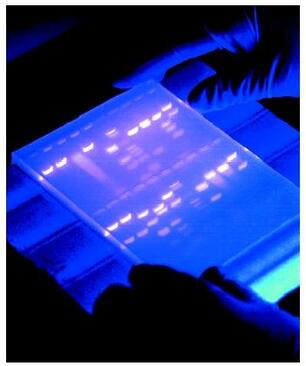
Agarose gel electrophoresis is a method used to separate DNA strands by size, and to determine the size of the separated strands by comparison to strands of known length. Your PCR products are deposited in the top of the gel. Using electricity, the DNA (with a negative charge) is pushed through the gel towards the positive electrode. As your gel "runs," the DNA is separated by size. The DNA strands show up as bands under UV light and you can read the results. Your products can be compared with the ladder or marker, which has standard sized DNA fragments of KNOWN length used for comparison. In this way, you can know the exact length of your DNA samples. -----> You will MAKE & RUN an agorose gel in Lab 11 to make sure our PCR product contains the cytochrome b gene. |
5) Sequencing
Once we know we have amplified (copied) the right gene we are ready to sequence the gene. We expect the sequence (the order of As, Ts, Cs and Gs) within the cytochrome b gene to be different for different species. Samples are placed into a sequencer apparatus which can detect the order of nucleotide bases in our sample. The sequence is then cleaned and edited, |
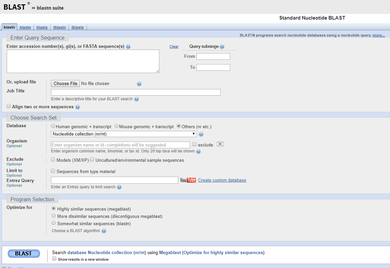
6) BLASTing
6) BLASTing: The National Institute of Health (NIH) and National Center for Biotechnology Information (NCBI) hosts a database called GenBank, which houses all known DNA sequences. Once the sequences of our samples are ready, they are pasted into a search tool (called a BLAST) which matches them to the correct species! -----> You will be provided the sequence of successful samples in Lab 11 and asked to determine the species of origin in lab. |
Lab 10: ProtocolYour task in Lab 10 is to replicate a DNA extraction protocol using fruit samples, to determine how much DNA we may have successfully extracted from our samples, and to learn about the technique of PCR.
Exercise I. Practice Extracting DNA Exercise II. Determine how much DNA we have. Exercise III. Virtual PCR |
-
Exercise I
-
Exercise II
-
Exercise III
<
>
Exercise I. How Does DNA Extraction Work?
Our meat samples have been collected, processed in the field and brought back to WKU. In the WKU Biotechnology Center, Ms. Naomi Rowland (our collaborator) has completed the digestion of the samples and a procedure designed to extract the DNA. DNA extraction is an important step in DNA analysis, which is at the forefront of biomedicine and forensics. DNA Extraction Buffer is used for this purpose.
Extraction of DNA from meat samples is a lengthy and difficult process. It has already been completed for our samples. Today, you will mirror this process: you will be extracting DNA from fruit samples. You may need to review the structure of DNA (covered in Ch 4 of your BIOL 120 Pearson text).
Extraction of DNA from meat samples is a lengthy and difficult process. It has already been completed for our samples. Today, you will mirror this process: you will be extracting DNA from fruit samples. You may need to review the structure of DNA (covered in Ch 4 of your BIOL 120 Pearson text).
DNA extraction includes the use of an extraction buffer and ethanol:
- Our extraction buffer includes detergent to break apart lipids and proteins and "free" the DNA from the nucleus (to get it out of the cell).
- It also includes salt with (+) charged sodium ions to neutralizes the (-) charge on the sugar-phosphate backbone of DNA, making it less soluble in water (so it stays together).
- We use ethanol to separate the DNA from the mixture. DNA is not soluble in ethanol so it is visible (so we can see it). For our purposes we will use isopropyl alcohol (rubbing alcohol) as a good substitute.
Materials.
|
Procedure.
|
Exercise II. What part of the DNA do we need?
Now, that understand the extraction process, let's move back to our bushmeat samples. Following extraction of DNA from the meat samples, we need to determine how much DNA we have. In other words...were we successful and to a high enough degree to allow for the next steps? Following extraction we have tons of DNA! But, that means that in fact we have a lot of genetic material that we do not need and certainly do not want to make copies of. DNA is conserved at different rates across species. Meaning we share different amounts and different genes. We need to find a gene (a section of DNA) that all mammals have, BUT that is a different enough in every species of mammal to afford us the ability to identify the species from the DNA sequence of the gene. The particular gene required is called the Gene of Interest or "GoI."
|
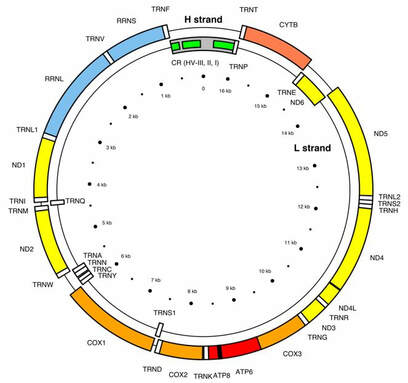
We will use the cytochrome b gene (CTYb) for species ID of our bushmeat samples. It meets our requirements. It is shared across all mammals BUT, differs enough between species for the sequence (of As, Ts, Cs and Gs) to give us a species-level ID. |
Procedure: Estimate the number of DNA fragments (our gene of interest, cytochrome b) we may have following extraction.
|
Exercise III. PCR Virtual Lab
Polymerase chain reaction (PCR) involves the amplification (or copying) of a specific segment or fragment of DNA to allow for continued analysis. PCR can be used to: identify individuals or species, for criminal proceedings, paternity determination or pathology. It is one of the most powerful tools of modern biology.
We would have created PCR cocktails for each of our bushmeat samples and placed into a thermocycler for synthesis. Instead, you will do a short virtual PCR lab. Read over the information in PCR below and complete the procedure.
|
Once DNA has been extracted, it is mixed into a particular PCR solution containing:
|
|
|
Lab 10 BIOL 120 CONNECTIONS Section 1.6: Doing Biology Big Picture 1: How to Think Like a Scientist Chapter 4: Nucleic Acids Chapter 9: Cellular Respiration Chapter 15: DNA and the Gene: Synthesis & Repair Chapter 16: How Genes Work Chapter 20: The Molecular Revolution Chapter 54: Biodiversity and Conservation Biology *BIOL 122 |
KAS citation format:
Mountjoy, N.J 2021. Title of page. Biological Concepts: Cells, Metabolism & Genetics. https://www.121cellmetagen.com. Date accessed (MM/DD/YYY).
Mountjoy, N.J 2021. Title of page. Biological Concepts: Cells, Metabolism & Genetics. https://www.121cellmetagen.com. Date accessed (MM/DD/YYY).